Research interests
Our research focuses on developing computational algorithms and conducting large-scale data mining of single-cell and spatial multi-omic data. We aim to unravel the intricate mechanisms of gene regulation and cellular crosstalk that define cell identities and their responses to environmental stimuli. We have created methods and web resources specifically designed for analyzing single-cell and spatial multi-omics data, including signal enhancement, multimodal generation, and network modeling. By employing AI to model single-cell spatial multimodal datasets, we seek to establish quantitative connections between changes in cell identity and disease phenotypes across various biological systems, including cancer, development, and aging.
Intelligent Algorithms for Enhancing, Generating, and Modeling Single-Cell Spatial Multi-Omics
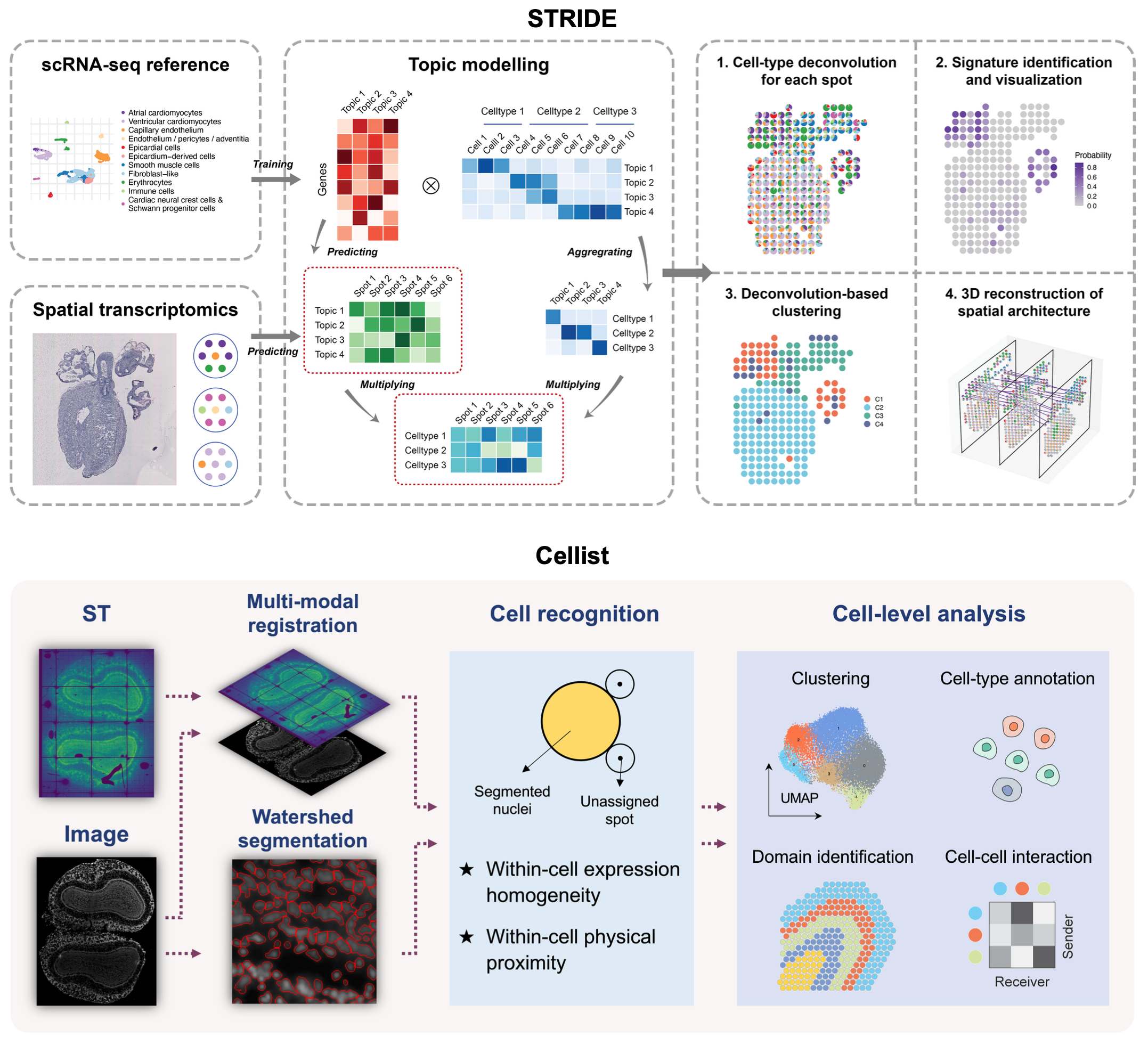
Single-cell and spatial multi-omic technologies have revolutionized our understanding of cellular heterogeneity in complex biological systems. However, analyses in this field currently face several challenges, including inadequate resolution, limited coverage, and difficulties in integrating and generating multimodal data. To address these issues, we have developed a series of intelligent algorithms. STRIDE deconvolutes low-resolution spatial transcriptomics data signals. Cellist segments and enhances high-resolution spatial transcriptomics data in to single-cells. SCRIP and SCRIPro construct gene regulation networks using single-cell and spatial multi-omics data. MetroSCREEN builds metabolic-based regulomes. EvaCCI assesses intercellular interactions. These algorithms enhance the usability of single-cell spatial omics data and lay the foundation for integrative analysis of spatiotemporal multimodal data, as well as computational modeling of cellular phenotypes in both physiological and pathological conditions.
Virtual Cell and Tissue Modeling Using Generative AI and Large-Scale Spatiotemporal Multimodal Data
Cell phenotypes in multicellular systems are regulated by intrinsic factors (such as gene expression) and extrinsic factors (such as intercellular interactions). We have demonstrated a strong connection between intrinsic epigenetic regulations and cell fate determination in mouse IVF and SCNT embryo development (Nature, 2016; Nat. Cell Biol., 2018; Cell Stem Cell, 2018, 2022; Cell Res., 2022). Currently, we are developing generative virtual cell and tissue models pretrained on large-scale single-cell spatial multimodal datasets. These models aim to uncover the collaborative effects of gene regulation, cellular crosstalk, and other environmental factors, such as metabolites and mechanical influences, on predicting cell and tissue phenotypes. We have developed SELINA, which utilizes a multi-adversarial domain adaptation network to automatically predict cell types using a large-scale pretrained human scRNA-seq reference. Our goal is to leverage generative AI models to gain deeper insights into the molecular mechanisms driving cell and tissue phenotypes, further guiding and reshaping this transformation process.
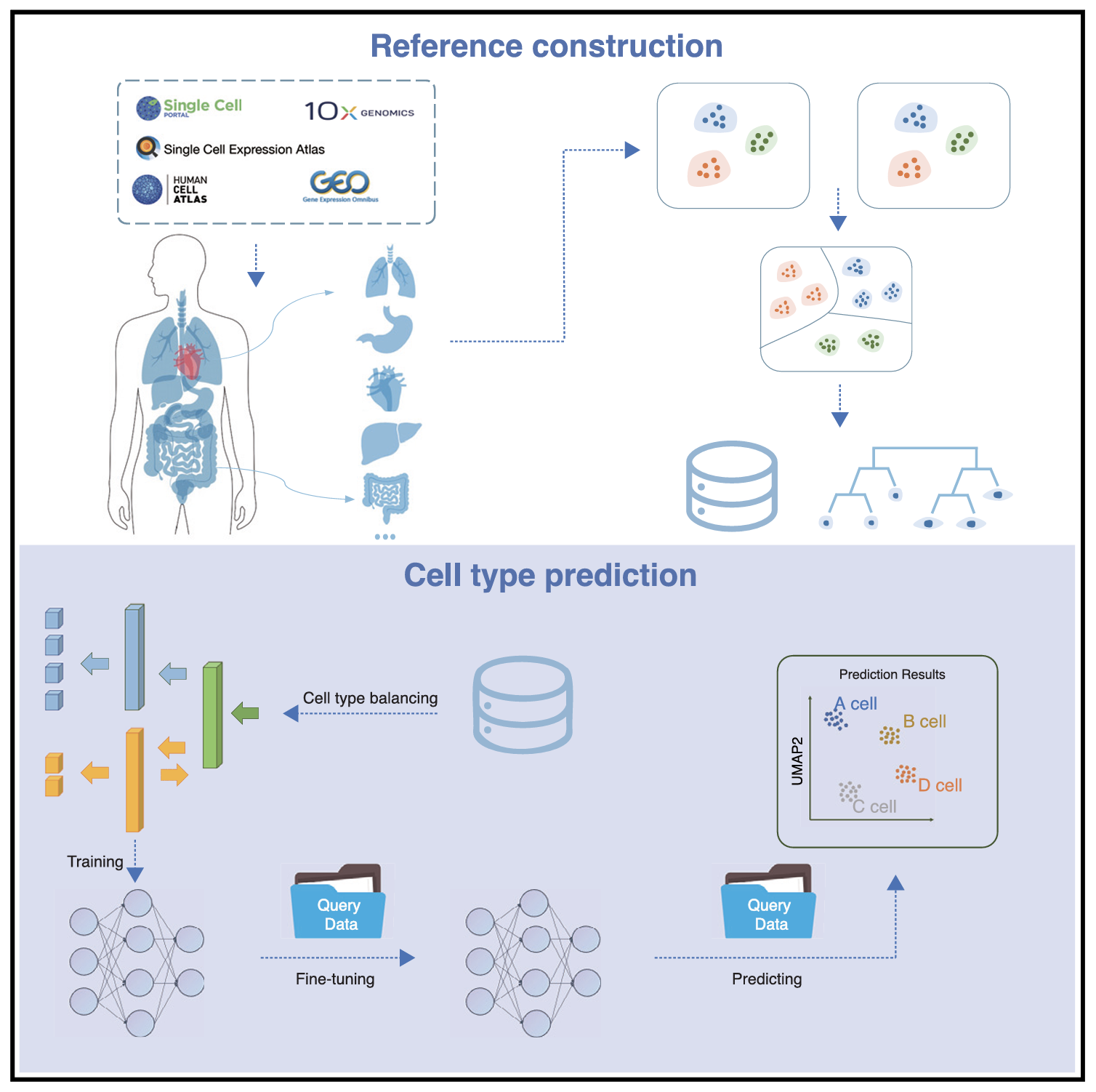
Connection of Cell Phenotypes to Individual Phenotypes in Complex Disease Microenvironments
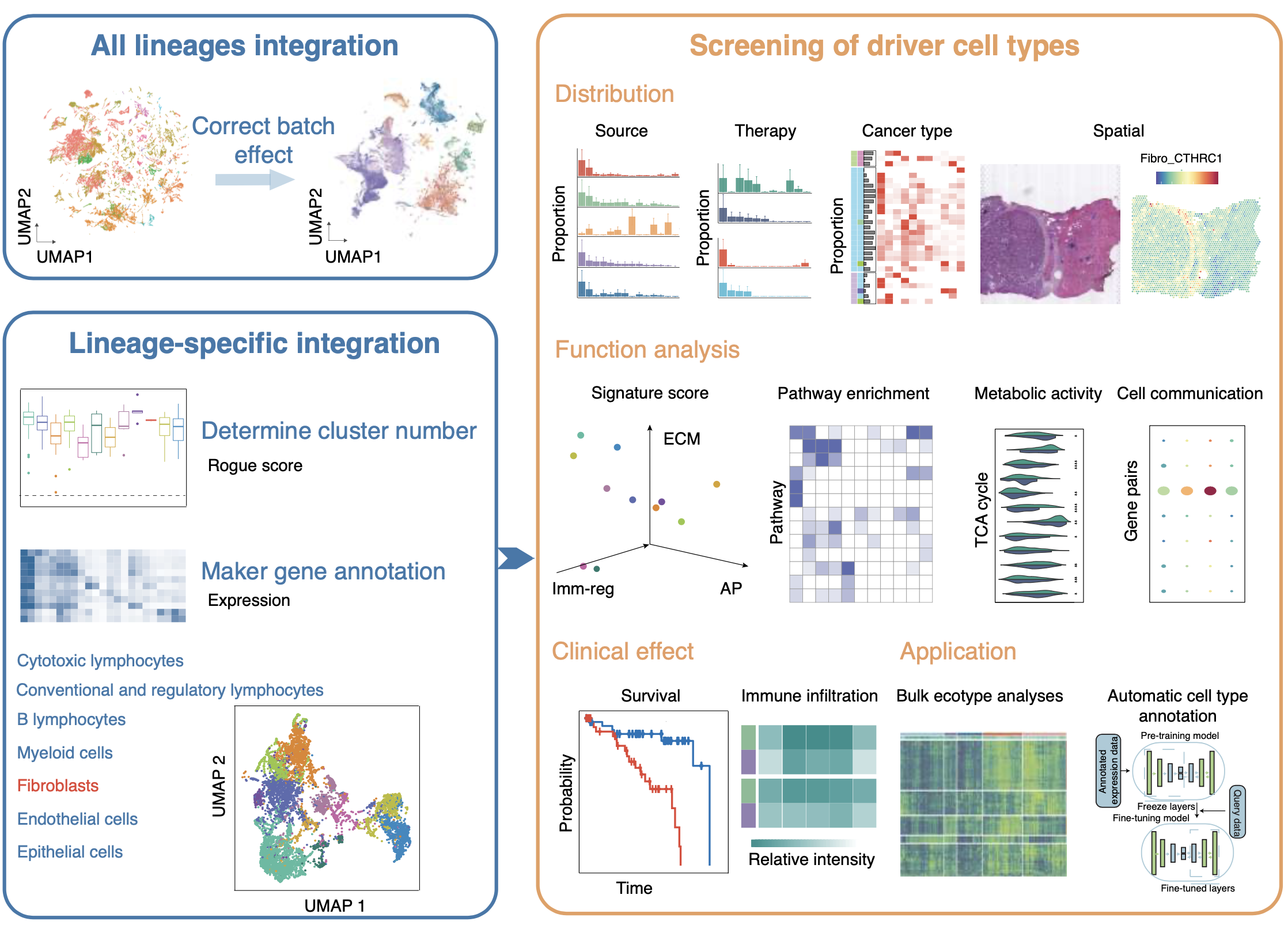
Cancer and aging-associated diseases result from an imbalance in cell and tissue phenotypes, shifting away from healthy states. Our team combines AI-driven virtual cell and tissue models with experimental validations to identify the drivers of disease-related cell and tissue phenotypes in tumors and aging-related diseases. We have established a comprehensive single-cell RNA sequencing data resource, TISCH, for analyzing gene expression and cellular composition in the tumor microenvironment (TME). Based on this resource, we constructed a pan-cancer cellular phenotype atlas called TabulaTIME, where we discovered a widely prevalent profibrotic ecotype that regulates tumor immunity (Nat. Cancer 2025). Currently, our team is collaborating closely with oncologists and immunologists to investigate the mechanisms of tumor microenvironment evolution and immune therapy resistance across different cancer types (Cell 2024; Nat. Genet. 2025; Cancer Immunol. Res. 2023; Genome Med. 2023; EMBO J. 2023).
